Applying Quality Checks
You can apply quality checks to your data using the following approaches:
- Programmatic approach: you can apply checks programmatically to a DataFrame or Tables. This method is required if you want to perform a data quality check for data in-transit.
- No-code approach (Workflows): you can apply checks for data at-rest using workflows if DQX is installed in the workspace as a tool. This does not require code-level integration and is suitable for data already stored in Delta tables or files.
DQX offers a set of predefined built-in quality rules (checks) as described here. All DQX rules are evaluated independently and in parallel. If one rule fails, the others are still processed. All rule types, including row-level and dataset-level rules, can be applied together in a single execution.
Programmatic approach
Checks can be applied to the input data by one of the following methods of the DQEngine class:
-
For checks defined with DQX classes:
apply_checks: apply quality checks to a DataFrame and report issues as additional columns.apply_checks_and_split: apply quality checks and split the input DataFrame into valid and invalid (quarantined) DataFrames.apply_checks_and_save_in_table: end-to-end approach to apply quality checks to a table and save results to the output table(s) via single method call.
-
For checks defined as metadata (list of dictionaries, or loaded from a storage):
apply_checks_by_metadata: apply quality checks to a DataFrame and report issues as additional columns.apply_checks_by_metadata_and_split: apply quality checks and split the input DataFrame into valid and invalid (quarantined) DataFrames.apply_checks_by_metadata_and_save_in_table: end-to-end approach to apply quality checks to a table and save results to output table(s) via single method call.
-
There are also method for applying checks to multiple input locations at once:
apply_checks_and_save_in_tables: apply quality checks to input locations defined by provided list ofRunConfig.apply_checks_and_save_in_tables_for_patterns: apply quality checks to tables matching wildcard patterns.
You can see the full list of DQEngine methods here.
The engine ensures that the specified column, columns, filter, or sql 'expression' fields can be resolved in the input DataFrame. If any of these fields are invalid, the check evaluation is skipped, and the results include the check failure with a message identifying the invalid fields.
The engine will raise an error if you try to apply checks with invalid definition (e.g. wrong syntax).
In addition, you can also perform a standalone syntax validation of the checks as described here.
You can apply quality checks to streaming pipelines using the same methods as for batch processing.
You can either use the end-to-end methods or manage the input stream and output directly with native Spark APIs (e.g. spark.readStream and writeStream).
You can find ready-to-use examples of applying checks with different methods in the demo section.
The results are reported as additional columns in the output DataFrame (by default _warning and _error columns).
See quality check results for details on the structure of the results.
You can customize the reporting columns as described in the additional configuration section.
Applying checks defined with DQX classes
- Python
from databricks.labs.dqx import check_funcs
from databricks.labs.dqx.engine import DQEngine
from databricks.labs.dqx.rule import DQRowRule, DQDatasetRule, DQForEachColRule
from databricks.labs.dqx.config import InputConfig, OutputConfig
from databricks.sdk import WorkspaceClient
dq_engine = DQEngine(WorkspaceClient())
checks = [
DQRowRule(
criticality="warn",
check_func=check_funcs.is_not_null,
column="col3",
),
DQDatasetRule(
criticality="error",
check_func=check_funcs.is_unique,
columns=["col1", "col2"],
),
DQRowRule(
name="email_invalid_format",
criticality="error",
check_func=check_funcs.regex_match,
column="email",
check_func_kwargs={"regex": "^[a-zA-Z0-9._%+-]+@[a-zA-Z0-9.-]+\.[a-zA-Z]{2,}$"},
),
]
input_df = spark.read.table("catalog.schema.input")
# Option 1: apply quality checks on the DataFrame and output results to a single DataFrame
valid_and_invalid_df = dq_engine.apply_checks(input_df, checks)
dq_engine.save_results_in_table(
output_df=valid_and_invalid_df,
output_config=OutputConfig(location="catalog.schema.output"),
)
# Option 2: apply quality checks on the DataFrame and provide valid and invalid (quarantined) DataFrames
valid_df, invalid_df = dq_engine.apply_checks_and_split(input_df, checks)
dq_engine.save_results_in_table(
output_df=valid_df,
quarantine_df=invalid_df,
output_config=OutputConfig(location="catalog.schema.valid"),
quarantine_config=OutputConfig(location="catalog.schema.quarantine"),
)
# Option 2b: save results to Unity Catalog Volume paths instead of tables
valid_df, invalid_df = dq_engine.apply_checks_and_split(input_df, checks)
dq_engine.save_results_in_table(
output_df=valid_df,
quarantine_df=invalid_df,
output_config=OutputConfig(location="/Volumes/catalog/schema/volume/valid_data", format="delta"),
quarantine_config=OutputConfig(location="/Volumes/catalog/schema/volume/quarantine_data", format="delta"),
)
# Option 3 End-to-End approach: apply quality checks to a table and save results to valid and invalid (quarantined) tables
dq_engine.apply_checks_and_save_in_table(
checks=checks,
input_config=InputConfig(location="catalog.schema.input"),
output_config=OutputConfig(location="catalog.schema.valid"),
quarantine_config=OutputConfig(location="catalog.schema.quarantine"),
)
# Option 4 End-to-End approach: apply quality checks to a table and save results to an output table
dq_engine.apply_checks_and_save_in_table(
checks=checks,
input_config=InputConfig(location="catalog.schema.input"),
output_config=OutputConfig(location="catalog.schema.output"),
)
Applying checks defined using metadata
Users can save and load checks as metadata (list of dictionaries) from a storage through several supported methods, as described here. When loading checks from a storage, they are always returned as metadata (list of dictionaries). You can convert checks from metadata to classes and back using serialization methods described here.
In the example below, checks are defined in YAML syntax for convenience and then loaded into a list of dictionaries.
- Python
from databricks.labs.dqx.engine import DQEngine
from databricks.labs.dqx.config import InputConfig, OutputConfig
from databricks.sdk import WorkspaceClient
dq_engine = DQEngine(WorkspaceClient())
# checks can also be loaded from a storage
checks: list[dict] = yaml.safe_load("""
- criticality: warn
check:
function: is_not_null
arguments:
column: col3
- criticality: error
check:
function: is_unique
arguments:
columns:
- col1
- col2
- name: email_invalid_format
criticality: error
check:
function: regex_match
arguments:
column: email
regex: ^[a-zA-Z0-9._%+-]+@[a-zA-Z0-9.-]+\.[a-zA-Z]{2,}$
""")
input_df = spark.read.table("catalog.schema.input")
# Option 1: apply quality checks on the DataFrame and output results as a single DataFrame
valid_and_invalid_df = dq_engine.apply_checks_by_metadata(input_df, checks)
dq_engine.save_results_in_table(
output_df=valid_and_invalid_df,
output_config=OutputConfig(location="catalog.schema.output"),
)
# Option 2: apply quality checks on the DataFrame and provide valid and invalid (quarantined) DataFrames
valid_df, invalid_df = dq_engine.apply_checks_by_metadata_and_split(input_df, checks)
dq_engine.save_results_in_table(
output_df=valid_df,
quarantine_df=invalid_df,
output_config=OutputConfig(location="catalog.schema.valid"),
quarantine_config=OutputConfig(location="catalog.schema.quarantine"),
)
# Option 3 End-to-End approach: apply quality checks on the input table and save results to valid and invalid (quarantined) tables
dq_engine.apply_checks_by_metadata_and_save_in_table(
checks=checks,
input_config=InputConfig(location="catalog.schema.input"),
output_config=OutputConfig(location="catalog.schema.valid"),
quarantine_config=OutputConfig(location="catalog.schema.quarantine"),
)
# Option 4 End-to-End approach: apply quality checks on the input table and save results to an output table
dq_engine.apply_checks_by_metadata_and_save_in_table(
checks=checks,
input_config=InputConfig(location="catalog.schema.input"),
output_config=OutputConfig(location="catalog.schema.output"),
)
Checks are applied to a column specified in the arguments of the check. This can be either a column name as string or column expression.
Some checks require columns (e.g. is_unique) instead of column, which is a list of column names or expressions. There are also checks that require no columns at all (e.g. sql_expression and sql_query).
Alternatively, for_each_column can be used to define a list of columns for which the same check should be applied. The for_each_column is supported for quality rules that take column or columns as arguments (e.g. not supported by sql_expression and sql_query).
For details on each check, please refer to the documentation here.
Applying checks on multiple tables
Applying checks on multiple tables can be performed by executing the same methods as described above in a loop.
However, DQX provides a convenient method to handle multiple tables in a single method call.
Use apply_checks_and_save_in_tables or apply_checks_and_save_in_tables_for_patterns to perform end-to-end quality checking for multiple tables.
Use explicit configuration for each table:
- Python
import yaml
from databricks.labs.dqx.config import InputConfig, OutputConfig, RunConfig
from databricks.labs.dqx.config import TableChecksStorageConfig
from databricks.labs.dqx.engine import DQEngine
from databricks.sdk import WorkspaceClient
dq_engine = DQEngine(WorkspaceClient())
# Define checks
checks = yaml.safe_load("""
- criticality: error
check:
function: is_not_null
arguments:
column: user_id
- criticality: warn
check:
function: regex_match
arguments:
column: email
regex: ^[a-zA-Z0-9._%+-]+@[a-zA-Z0-9.-]+\\.[a-zA-Z]{2,}$
""")
# Save checks in a table
checks_table = "catalog.schema.checks"
dq_engine.save_checks(checks, config=TableChecksStorageConfig(location=checks_table, run_config_name=checks_table, mode="overwrite"))
# Define configuration to check multiple tables
run_configs = [
RunConfig(
name=users_table,
input_config=InputConfig(location="catalog.schema.input_data_001"),
output_config=OutputConfig(
location="catalog.schema.output_data_001",
mode="overwrite"
),
# quarantine bad data
quarantine_config=OutputConfig(
location="catalog.schema.quarantine_data_001",
mode="overwrite"
),
checks_location=checks_table
# optional dictionary of reference DataFrames stored in tables/views to use in the checks
#reference_tables={"ref_df_key": InputConfig(location="catalog.ref_schema.reference")},
# optional mapping of fully qualified custom check function name to the module location
# (absolute or relative Workspace path or Volume path)
#custom_check_functions={"custom_check_func": "/Workspace/my_repo/my_module.py"},
),
RunConfig(
name=orders_table,
input_config=InputConfig(location="catalog.schema.input_data_002"),
# don't quarantine bad data
output_config=OutputConfig(
location="catalog.schema.output_data_002",
mode="overwrite"
),
checks_location=checks_table
)
]
# Apply checks
dq_engine.apply_checks_and_save_in_tables(run_configs=run_configs)
Apply checks on tables using wildcard patterns:
- Python
import yaml
from databricks.labs.dqx.config import InputConfig, OutputConfig, RunConfig
from databricks.labs.dqx.profiler.profiler import DQProfiler
from databricks.labs.dqx.profiler.generator import DQGenerator
from databricks.labs.dqx.config import TableChecksStorageConfig
from databricks.labs.dqx.engine import DQEngine
from databricks.sdk import WorkspaceClient
dq_engine = DQEngine(WorkspaceClient())
# Perform quality checking on all tables matching the patterns
patterns = ["catalog.schema.*", "catalog.schema2.input_data_003"]
# Skip output tables using suffixes
exclude_patterns=["*_dq_output", "*_dq_quarantine", "*_checked", "*_quarantine"]
# (Optional step) profile input data and generate quality rules candidates
profiler = DQProfiler(ws, spark)
generator = DQGenerator(ws)
# Profile tables based on patterns and
results = profiler.profile_tables_for_patterns(
patterns=patterns,
exclude_patterns=exclude_patterns,
)
# Save quality checks to a table
for table, (summary_stats, profiles) in results.items():
checks = generator.generate_dq_rules(profiles)
# run config name must be equal to the input table name
dq_engine.save_checks(checks, config=TableChecksStorageConfig(location=checks_table, run_config_name=table, mode="overwrite"))
# Apply checks on multiple tables using patterns
dq_engine.apply_checks_and_save_in_tables_for_patterns(
patterns=patterns,
checks_location=checks_table, # run config of the saved checks name must be equal to the input table name
)
# Apply checks on multiple tables using patterns and prefilled run config parameters
dq_engine.apply_checks_and_save_in_tables_for_patterns(
patterns=patterns,
exclude_patterns=exclude_patterns,
checks_location=checks_table,
run_config_template=RunConfig(
# auto-created if not provided; location skipped and derived from patterns
input_config=InputConfig(""),
# auto-created if not provided; location skipped and derived from patterns + output_table_suffix
output_config=OutputConfig(location="", mode="overwrite"),
# quarantine bad data; location skipped and derived from patterns + quarantine_table_suffix
quarantine_config=OutputConfig(location="", mode="overwrite"),
# skip checks_location of the run config as it is derived separately
# optional dictionary of reference DataFrames stored in tables/views to use in the checks
#reference_tables={"ref_df_key": InputConfig(location="catalog.ref_schema.reference")},
# optional mapping of fully qualified custom check function name to the module location
# (absolute or relative Workspace path or Volume path)
#custom_check_functions={"custom_check_func": "/Workspace/my_repo/my_module.py"},
),
output_table_suffix="_checked", # default _dq_output
quarantine_table_suffix="_quarantine" # default _dq_quarantine
)
Applying checks in Lakeflow Pipelines
Lakeflow Pipelines (formerly DLT - Delta Live Tables) provides expectations to enforce data quality constraints. However, expectations don't offer detailed insights into why certain checks fail and don't provide native quarantine functionality.
The example below demonstrates integrating DQX with Lakeflow Pipelines to provide comprehensive quality information. The DQX integration with Lakeflow Pipelines does not use Expectations but DQX's own methods.
You can define checks using DQX classes or as metadata (list of dictionaries) or loaded from YAML/JSON file or a table as described here.
Option 1: Apply quality check and quarantine bad records
- Python
import dlt
from databricks.labs.dqx.engine import DQEngine
from databricks.sdk import WorkspaceClient
dq_engine = DQEngine(WorkspaceClient())
checks = ... # quality rules / checks defined as metadata or DQX classes
@dlt.view
def bronze_dq_check():
df = dlt.read_stream("bronze")
return dq_engine.apply_checks_by_metadata(df, checks)
@dlt.table
def silver():
df = dlt.read_stream("bronze_dq_check")
# get rows without errors or warnings, and drop auxiliary columns
return dq_engine.get_valid(df)
@dlt.table
def quarantine():
df = dlt.read_stream("bronze_dq_check")
# get only rows with errors or warnings
return dq_engine.get_invalid(df)
Option 2: Apply quality checks and report issues as additional columns
- Python
import dlt
from databricks.labs.dqx.engine import DQEngine
from databricks.sdk import WorkspaceClient
checks = ... # quality rules / checks defined as metadata or DQX classes
dq_engine = DQEngine(WorkspaceClient())
@dlt.view
def bronze_dq_check():
df = dlt.read_stream("bronze")
return dq_engine.apply_checks_by_metadata(df, checks)
@dlt.table
def silver():
df = dlt.read_stream("bronze_dq_check")
return df
Applying checks using foreachBatch
Spark's foreachBatch allows users to define an arbitrary processing function run for each streaming batch. DQX requires authentication with a Databricks SDK WorkspaceClient which is not supported inside foreachBatch. To use DQX in a foreachBatch function, create a DQEngine instance with a mock WorkspaceClient.
Some DQX engine methods require a valid Databricks WorkspaceClient. Methods must support local execution to be run in a foreachBatch function with a mocked WorkspaceClient. Check the Supports local execution column here to verify that DQX engine methods can be executed with a mocked client.
from unittest.mock import MagicMock
from pyspark.sql import DataFrame
from databricks.labs.dqx import check_funcs
from databricks.labs.dqx.engine import DQEngine
from databricks.labs.dqx.rule import DQRowRule
from databricks.sdk import WorkspaceClient
# Define DQX checks
checks = [
DQRowRule(
name="col_timestamp_is_null",
criticality="error",
check_func=check_funcs.is_not_null,
column="timestamp"
),
DQRowRule(
name="col_value_is_null",
criticality="error",
check_func=check_funcs.is_not_null,
column="value"
)
]
# Define a `foreachBatch` function to check and write the data
def validate_and_write_micro_batch_data(batch_df: DataFrame, _: int) -> None:
# Create a mock `WorkspaceClient`
mock_ws = MagicMock(spec=WorkspaceClient)
# Create the `DQEngine`
dq_engine = DQEngine(mock_ws)
# Apply the DQX checks and write the data
valid_and_invalid_df = dq_engine.apply_checks(batch_df, checks)
valid_and_invalid_df.write.format("delta").saveAsTable("catalog.schema.output")
# Read the data and apply the `foreachBatch` function
input_data = spark.readStream.format("rate-micro-batch").option("rowsPerBatch", 100).load()
input_data.writeStream.foreachBatch(validate_and_write_micro_batch_data).start()
Saving quality results
The quality check results (DataFrames) can be saved by the user to arbitrary tables using Spark APIs. DQX also offers convenient methods to save the results to tables without the need to use Spark APIs directly. Users can also use end-to-end methods that handle applying checks and saving results in a single method.
You can see the full list of DQEngine methods here.
Below are examples on how to save the quality checking results in tables using the DQEngine methods:
from databricks.labs.dqx.config import OutputConfig
from databricks.labs.dqx.engine import DQEngine
from databricks.sdk import WorkspaceClient
dq_engine = DQEngine(WorkspaceClient())
# apply checks
valid_df, invalid_df = dq_engine.apply_checks_and_split(input_df, checks)
# use spark native APIs to save the results
valid_df.write.options({"mergeSchema": "true"}).mode("append").saveAsTable("catalog.schema.valid")
invalid_df.write.options({"mergeSchema": "true"}).mode("append").saveAsTable("catalog.schema.quarantine")
# save results to tables using DQX method
dq_engine.save_results_in_table(
output_df=valid_df,
quarantine_df=invalid_df,
output_config=OutputConfig(
location="catalog.schema.valid",
mode="append", # or "overwrite"
options={"mergeSchema": "true"}, # default {}
),
quarantine_config=OutputConfig(
location="catalog.schema.quarantine",
mode="append", # or "overwrite"
options={"mergeSchema": "true"}, # default {}
),
)
# save results to tables using streaming using DQX method
dq_engine.save_results_in_table(
output_df=valid_df,
quarantine_df=invalid_df, # optional
output_config=OutputConfig(
location="catalog.schema.valid",
trigger={"availableNow": True},
options={"checkpointLocation": "/path/to/checkpoint_output", "mergeSchema": "true"},
),
# required if quarantine_df is defined
quarantine_config=OutputConfig(
location="catalog.schema.quarantine",
trigger={"availableNow": True},
options={"checkpointLocation": "/path/to/checkpoint_quarantine", "mergeSchema": "true"},
),
)
# save results to tables defined in run config using DQX method
dq_engine.save_results_in_table(
output_df=valid_df,
quarantine_df=invalid_df,
run_config_name="default", # use run config to get output and quarantine table details, and writing options
)
# End-to-end method: apply checks and save results in tables using DQX method
dq_engine.apply_checks_and_save_in_table(
checks=checks,
input_config=InputConfig(
location="catalog.schema.input"
),
output_config=OutputConfig(
location="catalog.schema.valid",
mode="append", # or "overwrite"
),
# optional
quarantine_config=OutputConfig(
location="catalog.schema.quarantine",
mode="append", # or "overwrite"
),
)
# End-to-end method: apply checks and save results in tables using streaming using DQX method
dq_engine.apply_checks_and_save_in_table(
checks=checks,
input_config=InputConfig(
location="catalog.schema.input",
is_streaming=True,
),
output_config=OutputConfig(
location="catalog.schema.valid",
mode="append", # or "update" or "complete"
trigger={"availableNow": True},
options={"checkpointLocation": "/path/to/checkpoint_output", "mergeSchema": "true"},
),
# optional
quarantine_config=OutputConfig(
location="catalog.schema.quarantine",
mode="append", # or "update" or "complete"
trigger={"availableNow": True},
options={"checkpointLocation": "/path/to/checkpoint_quarantine", "mergeSchema": "true"},
),
)
No-code approach (Workflows)
You can apply quality checks to data at-rest using DQX Workflows without writing any code. This requires installation of DQX in the workspace as a tool (see installation guide).
The workflows use configuration file to specify input and output locations, and to apply sets of quality checks from a storage.
You can open the configuration file (.dqx/config.yml) in the installation folder from Databricks UI or by executing the following Databricks CLI command:
databricks labs dqx open-remote-config
There are currently two workflows available in DQX: quality checker and end-to-end workflow.
The workflows can be run manually or scheduled to run periodically. The workflows are not scheduled to run automatically by default minimizing concerns regarding compute and associated costs.
The workflows can be executed as batch or streaming jobs.
Quality Checker Workflow
Quality checker workflow performs the following:
- apply checks to the input data (files or a table)
- save results to output tables (with or without a quarantine)
To run the workflows manually, you can use Databricks UI or Databricks CLI:
# Run for all run configs in the configuration file
databricks labs dqx apply-checks
# Run for a specific run config in the configuration file
databricks labs dqx apply-checks --run-config "default"
# Run for tables/views matching wildcard patterns. Conventions:
# * Run config from configuration file is used as a template for all relevant fields except location
# * Input table location is derived from the patterns
# * For table-based checks location, checks are loaded from the specified table
# * For file-based checks location, file in the path is replaced with <input_table>.yml. In addition, if the location is specified as a relative path, it is prefixed with the installation folder
# * For output and quarantine tables location, use <input_table>_dq_output and <input_table>_dq_quarantine suffixes
# * By default, output and quarantine tables are excluded based on suffixes
# * Default for output table suffix is "_dq_output"
# * Default for quarantine table suffix is "_dq_quarantine"
databricks labs dqx apply-checks --run-config "default" --patterns "main.product001.*;main.product002"
# Run for wildcard patterns, exclude patterns, and custom output, and quarantine table suffixes
databricks labs dqx apply-checks --run-config "default" --patterns "main.product001.*;main.product002" --exclude-patterns "*_dq_output;*_dq_quarantine" --output-table-suffix "_output" --quarantine-table-suffix "_quarantine"
You can also provide --timeout-minutes option.
When running the quality checker workflow using Databricks API or UI, you have the same execution options:
- By default, the workflow executes across all defined run configs within the configuration file.
- To execute the workflow for a specific run config, specify the
run_config_nameparameter during execution. - To execute the workflow for tables/views matching wildcard patterns, provide value for
run_config_nameandpatternsparameters during execution. The following parameters are supported for pattern based execution:patterns: Accepts a semicolon-delimited list of patterns, e.g., "catalog.schema1.*;catalog.schema2.table1".run_config_name: Specifies the run config to use as a template for all relevant settings, except location field ('input_config.location') which is derived from the patterns.- The output and quarantine table names are derived by appending the value of
output_table_suffixandquarantine_table_suffixjob parameters, respectively, to the input table name. - If the
checks_locationin the run config points to a table, the checks will be directly loaded from that table. If thechecks_locationin the run config points to a file, file name is replaced with "<input_table>.yml". In addition, if the location is specified as a relative path, it is prefixed with the workspace installation folder. For example:- If "checks_location=catalog.schema.table", the location will be resolved to "catalog.schema.table" or "database.schema.table" in case of using Lakebase to store checks.
- If "checks_location=folder/checks.yml", the location will be resolved to "install_folder/folder/<input_table>.yml".
- If "checks_location=/App/checks.yml", the location will be resolved to "/App/<input_table>.yml".
- If "checks_location=/Volume/catalog/schema/folder/checks.yml", the location will be resolved to "/Volume/catalog/schema/folder/<input_table>.yml".
exclude_patterns: (optional) Accepts a semicolon-delimited list of patterns to exclude, e.g., "*_output;*_quarantine".- By default, the workflow skips output and quarantine tables automatically based on the
output_table_suffixandquarantine_table_suffixjob parameters. This prevents applying checks to the output and quarantine tables from previous runs. output_table_suffix: (optional) Suffix to append to the output table names (default: "_dq_output").quarantine_table_suffix: (optional) Suffix to append to the quarantine table names (default: "_dq_quarantine").
See the relevant params in red:
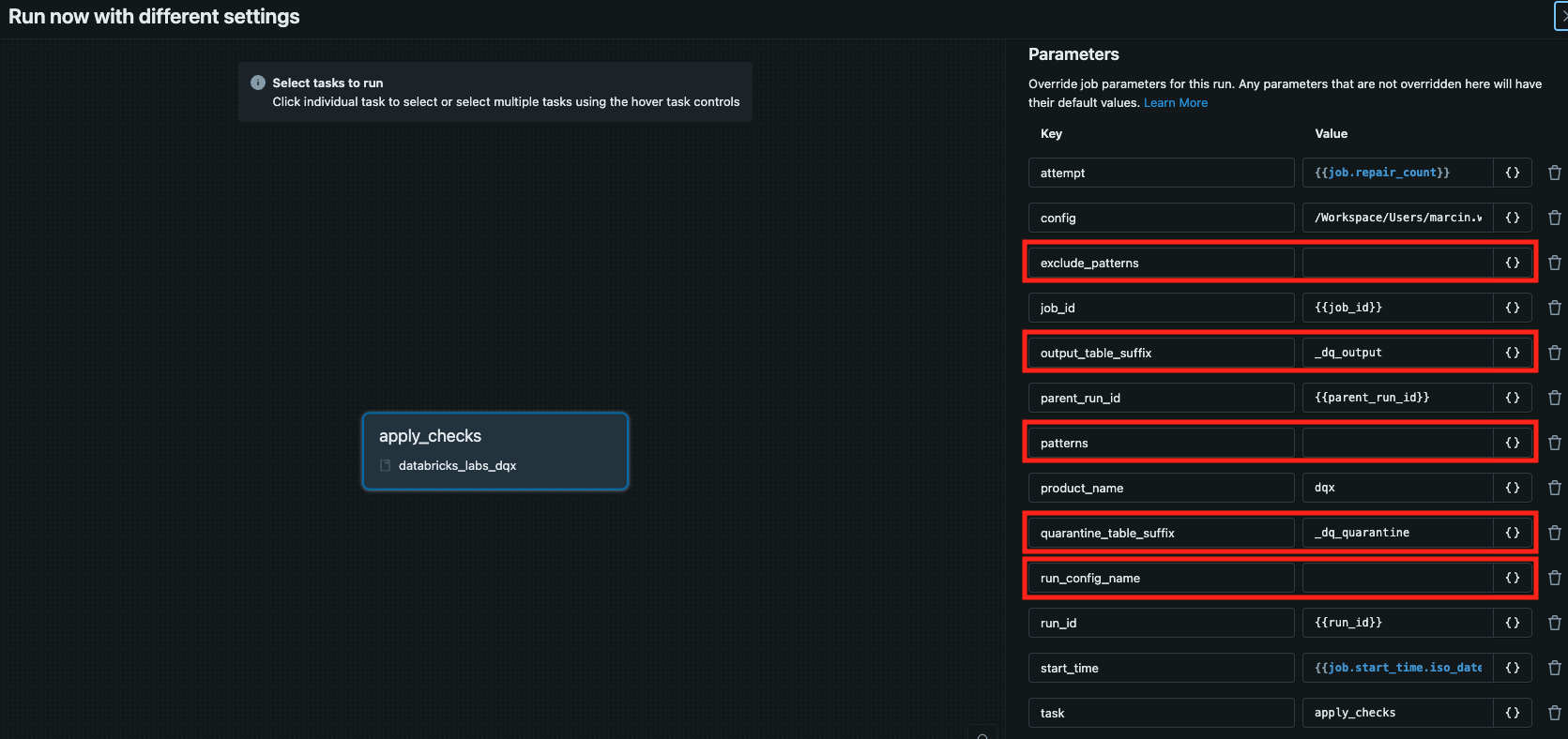
Execution logs are printed in the console and saved in the installation folder. You can display the logs from the latest run by executing the following command:
databricks labs dqx logs --workflow quality-checker
The following fields from the configuration file are used by the quality checker workflow:
input_config: configuration for the input data. 'location' is autogenerated when the workflow is executed for patterns.output_config: configuration for the output data. 'location' is autogenerated when the workflow is executed for patterns.quarantine_config: (optional) configuration for the quarantine data. 'location' is autogenerated when the workflow is executed for patterns.checks_location: location of the quality checks in storage. Autogenerated when the workflow is executed for patterns.serverless_clusters: whether to use serverless clusters for running the workflow (default:true). Using serverless clusters is recommended as it allows for automated cluster management and scaling.quality_checker_spark_conf: (optional) spark configuration to use for the workflow, only applicable ifserverless_clustersis set tofalse.quality_checker_override_clusters: optional cluster configuration to use for the workflow, only applicable ifserverless_clustersis set tofalse.quality_checker_max_parallelism: (optional) max parallelism for quality checking multiple tables (default: 4).extra_params: (optional) extra parameters to pass to the jobs such as result column names and user_metadata.custom_check_functions: (optional) mapping of custom check function name to Python file (module) containing check function definition.reference_tables: (optional) mapping of reference table names to reference table locations.
Example of the configuration file (relevant fields only):
serverless_clusters: true # default is true, set to false to use standard clusters
quality_checker_max_parallelism: 4 # <- max parallelism for quality checking multiple tables
run_configs:
- name: default
checks_location: catalog.table.checks # can be a table or file
input_config:
format: delta
location: /databricks-datasets/delta-sharing/samples/nyctaxi_2019
is_streaming: false # set to true if wanting to run it using streaming
output_config:
format: delta
location: main.nytaxi.output
mode: append
#checkpointLocation: /Volumes/catalog/schema/volume/checkpoint # only applicable if input_config.is_streaming is enabled
#trigger: # streaming trigger, only applicable if input_config.is_streaming is enabled
# availableNow: true
quarantine_config: # optional
format: delta
location: main.nytaxi.quarantine
mode: append
#checkpointLocation: /Volumes/catalog/schema/volume/checkpoint # only applicable if input_config.is_streaming is enabled
#trigger: # streaming trigger, only applicable if input_config.is_streaming is enabled
# availableNow: true
custom_check_functions: # optional
my_func: custom_checks/my_funcs.py # relative workspace path (installation folder prefix applied)
my_other: /Workspace/Shared/MyApp/my_funcs.py # or absolute workspace path
email_mask: /Volumes/main/dqx_utils/custom/email.py # or UC Volume path
reference_tables: # optional
reference_vendor:
input_config:
format: delta
location: main.nytaxi.ref
End-to-End Workflow
The end-to-end (e2e) workflow is designed to automate the entire data quality checking process, from profiling the input data to applying quality checks and saving results. The workflow executes the profiler and quality checker workflows in sequence, allowing you to generate quality checks based on the input data and then apply those checks to the same data.
End-to-end (e2e) workflow performs the following:
- profile input data (files or a table)
- generate quality checks
- apply the generated checks to the input data
- save results to output tables (with or without a quarantine)
To run the workflows manually, you can use Databricks UI or Databricks CLI:
# Run for all run configs in the configuration file
databricks labs dqx e2e
# Run for a specific run config in the configuration file
databricks labs dqx e2e --run-config "default"
# Run for tables/views matching wildcard patterns. Conventions:
# * Run config from configuration file is used as a template for all relevant fields except location
# * Input table location is derived from the patterns
# * For table-based checks location, checks are saved/loaded from the specified table
# * For file-based checks location, file in the path is replaced with <input_table>.yml. In addition, if the location is specified as a relative path, it is prefixed with the installation folder
# * For output and quarantine tables location, use <input_table>_dq_output and <input_table>_dq_quarantine suffixes
# * By default, output and quarantine tables are excluded based on suffixes
# * Default for output table suffix is "_dq_output"
# * Default for quarantine table suffix is "_dq_quarantine"
databricks labs dqx e2e --run-config "default" --patterns "main.product001.*;main.product002"
# Run for wildcard patterns, exclude patterns, and custom output and quarantine table suffixes
databricks labs dqx e2e --run-config "default" --patterns "main.product001.*;main.product002" --exclude-patterns "*_dq_output;*_dq_quarantine" --output-table-suffix "_output" --quarantine-table-suffix "_quarantine"
You can also provide --timeout-minutes option.
When running the e2e workflow using Databricks API or UI, you have the same execution options:
- By default, the workflow executes across all defined run configs within the configuration file.
- To execute the workflow for a specific run config, specify the
run_config_nameparameter during execution. - To execute the workflow for tables/views matching wildcard patterns, provide value for
run_config_nameandpatternsparameters during execution. The following parameters are supported for pattern based execution:patterns: Accepts a semicolon-delimited list of patterns, e.g., "catalog.schema1.*;catalog.schema2.table1".run_config_name: Specifies the run config to use as a template for all relevant settings, except location field ('input_config.location') which is derived from the patterns.- The output and quarantine table names are derived by appending the value of
output_table_suffixandquarantine_table_suffixjob parameters, respectively, to the input table name. - If the
checks_locationin the run config points to a table, the checks will be directly loaded from that table. If thechecks_locationin the run config points to a file, file name is replaced with <input_table>.yml. In addition, if the location is specified as a relative path, it is prefixed with the workspace installation folder. For example:- If "checks_location=catalog.schema.table", the location will be resolved to "catalog.schema.table" or "database.schema.table" in case of using Lakebase to store checks.
- If "checks_location=folder/checks.yml", the location will be resolved to "install_folder/folder/<input_table>.yml".
- If "checks_location=/App/checks.yml", the location will be resolved to "/App/<input_table>.yml".
- If "checks_location=/Volume/catalog/schema/folder/checks.yml", the location will be resolved to "/Volume/catalog/schema/folder/<input_table>.yml".
exclude_patterns: (optional) Accepts a semicolon-delimited list of patterns to exclude, e.g., "*_output;*_quarantine".- By default, the workflow skips output and quarantine tables automatically based on the
output_table_suffixandquarantine_table_suffixjob parameters. This prevents applying checks to the output and quarantine tables from previous runs. output_table_suffix: (optional) Suffix to append to the output table names (default: "_dq_output").quarantine_table_suffix: (optional) Suffix to append to the quarantine table names (default: "_dq_quarantine").
See the relevant params in red:
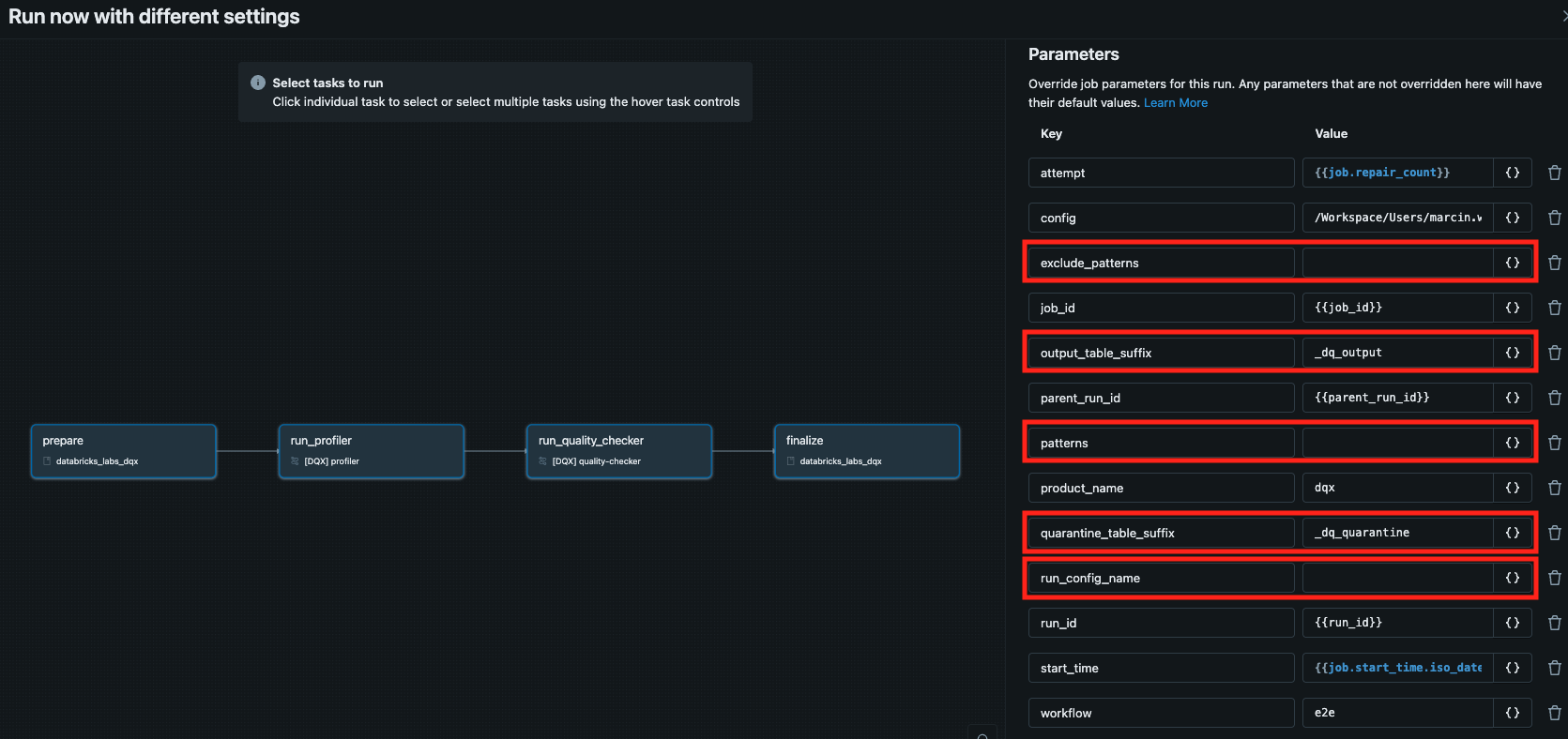
Execution logs are printed in the console and saved in the installation folder. You can display the logs from the latest run by executing the following command:
databricks labs dqx logs --workflow e2e
# see logs of the profiler and quality-checker jobs triggered by the e2e workflow
databricks labs dqx logs --workflow profiler
databricks labs dqx logs --workflow quality-checker
The following fields from the configuration file are used by e2e workflow:
input_config: configuration for the input data. 'location' is autogenerated when the workflow is executed for patterns.output_config: configuration for the output data. 'location' is autogenerated when the workflow is executed for patterns.quarantine_config: (optional) configuration for the quarantine data. 'location' is autogenerated when the workflow is executed for patterns.checks_location: location of the quality checks in storage. Autogenerated when the workflow is executed for patterns.metrics_config: (optional) configuration for storing summary metrics.serverless_clusters: whether to use serverless clusters for running the workflow (default:true). Using serverless clusters is recommended as it allows for automated cluster management and scaling.e2e_spark_conf: (optional) spark configuration to use for the e2e workflow, only applicable ifserverless_clustersis set tofalse.e2e_override_clusters: (optional) cluster configuration to use for the e2e workflow, only applicable ifserverless_clustersis set tofalse.quality_checker_spark_conf: (optional) spark configuration to use for the quality checker workflow, only applicable ifserverless_clustersis set tofalse.quality_checker_override_clusters: (optional) cluster configuration to use for the quality checker workflow, only applicable ifserverless_clustersis set tofalse.quality_checker_max_parallelism: (optional) max parallelism for quality checking multiple tables (default: 4).profiler_spark_conf: (optional) spark configuration to use with the profiler workflow, only applicable ifserverless_clustersis set tofalse.profiler_override_clusters: (optional) cluster configuration to use for profiler workflow, only applicable ifserverless_clustersis set tofalse.profiler_max_parallelism: (optional) max parallelism for profiling multiple tables (default: 4).profiler_config: configuration for the profiler containing:summary_stats_file: relative location within the installation folder of the summary statistics (default:profile_summary.yml)sample_fraction: fraction of data to sample for profiling.sample_seed: seed for reproducible sampling.limit: maximum number of records to analyze.filter: filter for the input data as a string SQL expression (default: None).
checks_user_requirements: (optional) user input for AI-assisted rule generationllm_config: (optional) configuration for the AI-assisted featuresextra_params: (optional) extra parameters to pass to the jobs such as result column names and user_metadatacustom_metrics: (optional) list of Spark SQL expressions for capturing custom summary metrics. By default, the number of input, warning, and error rows will be tracked. When custom metrics are defined, they will be tracked in addition to the default metrics.custom_check_functions: (optional) custom check functions defined in Python files that can be used in the quality checks.reference_tables: (optional) reference tables that can be used in the quality checks.
Example of the configuration file (relevant fields only):
serverless_clusters: true # default is true, set to false to use standard clusters
profiler_max_parallelism: 4 # max parallelism for profiling multiple tables
quality_checker_max_parallelism: 4 # max parallelism for quality checking multiple tables
llm_config: # configuration for the llm-assisted features
model:
model_name: "databricks/databricks-claude-sonnet-4-5"
api_key: xxx # optional API key for the model as secret in the format: secret_scope/secret_key. Not required by foundational models
api_base: xxx # optional API base for the model as secret in the format: secret_scope/secret_key. Not required by foundational models
run_configs:
- name: default
checks_location: catalog.table.checks # can be a table or file
input_config:
format: delta
location: /databricks-datasets/delta-sharing/samples/nyctaxi_2019
is_streaming: false # set to true if wanting to run it using streaming
output_config:
format: delta
location: main.nytaxi.output
mode: append
#checkpointLocation: /Volumes/catalog/schema/volume/checkpoint # only applicable if input_config.is_streaming is enabled
#trigger: # streaming trigger, only applicable if input_config.is_streaming is enabled
# availableNow: true
quarantine_config: # optional
format: delta
location: main.nytaxi.quarantine
mode: append
#checkpointLocation: /Volumes/catalog/schema/volume/checkpoint # only applicable if input_config.is_streaming is enabled
#trigger: # streaming trigger, only applicable if input_config.is_streaming is enabled
# availableNow: true
metrics_config: # optional - summary metrics storage
format: delta
location: main.nytaxi.dq_metrics
mode: append
profiler_config:
limit: 1000
sample_fraction: 0.3
summary_stats_file: profile_summary_stats.yml
llm_primary_key_detection: false
checks_user_requirements: "business rules description" # optional user input for the llm-assisted features
custom_check_functions: # optional
my_func: custom_checks/my_funcs.py # relative workspace path (installation folder prefix applied)
my_other: /Workspace/Shared/MyApp/my_funcs.py # or absolute workspace path
email_mask: /Volumes/main/dqx_utils/custom/email.py # or UC Volume path
reference_tables: # optional
reference_vendor:
input_config:
format: delta
location: main.nytaxi.ref
# Global custom metrics for summary statistics (optional)
custom_metrics:
- "sum(array_size(_warnings)) as total_warnings"
- "sum(array_size(_errors)) as total_errors"
Summary Metrics
DQX can capture and store summary metrics for your data quality checks in a centralized Delta table. These metrics can complement row-level quality checks stored in output DataFrames or be stored independently.
When enabled, the system collects both default summary metrics (input row count, error row count, warning row count, valid row count) and any custom summary metrics you define. Metrics can be configured programmatically or via a configuration file when installing DQX as a tool in the workspace.
Enabling summary metrics programmatically
To enable summary metrics programmatically, create and pass a DQMetricsObserver when initializing the DQEngine:
- Python
from databricks.labs.dqx.engine import DQEngine
from databricks.labs.dqx.metrics_observer import DQMetricsObserver
from databricks.sdk import WorkspaceClient
# Create the engine with the optional observer
dq_observer = DQMetricsObserver()
dq_engine = DQEngine(WorkspaceClient(), observer=dq_observer)
# Option 1: apply quality checks, provide a single result DataFrame, and return a metrics observation
valid_and_invalid_df, metrics_observation = dq_engine.apply_checks(input_df, checks)
valid_and_invalid_df.count() # Trigger an action to populate metrics (e.g. count, save to a table), otherwise accessing them will result in a stall
print(metrics_observation.get)
# save the metrics to a table
engine.save_summary_metrics(observed_metrics=metrics_observation.get, metrics_config=OutputConfig(location="catalog.schema.metrics"))
# Option 2: apply quality checks on the DataFrame, provide valid and invalid (quarantined) DataFrames, and return a metrics observation
valid_df, invalid_df, metrics_observation = dq_engine.apply_checks_and_split(input_df, checks)
invalid_df.count() # Trigger an action to populate metrics (e.g. count, save to a table), otherwise accessing them will result in a stall
print(metrics_observation.get)
# save the metrics to a table
engine.save_summary_metrics(observed_metrics=metrics_observation.get, metrics_config=OutputConfig(location="catalog.schema.metrics"))
# Option 3 End-to-End approach: apply quality checks to a table, save results to valid and invalid (quarantined) tables, and save metrics to a metrics table
dq_engine.apply_checks_and_save_in_table(
checks=checks,
input_config=InputConfig(location="catalog.schema.input"),
output_config=OutputConfig(location="catalog.schema.valid"),
quarantine_config=OutputConfig(location="catalog.schema.quarantine"),
metrics_config=OutputConfig(location="catalog.schema.metrics"),
)
# Option 4 End-to-End approach: apply quality checks to a table, save results to an output table, and save metrics to a metrics table
dq_engine.apply_checks_and_save_in_table(
checks=checks,
input_config=InputConfig(location="catalog.schema.input"),
output_config=OutputConfig(location="catalog.schema.output"),
metrics_config=OutputConfig(location="catalog.schema.metrics"),
)
Enabling summary metrics in DQX workflows
Summary metrics can also be enabled in DQX workflows. Metrics are configured:
- During installation for default run config: When prompted, choose to store summary metrics and configure the default metrics table location.
- Configuration file: Add
custom_metricsandmetrics_configto the configuration file.
For detailed information about summary metrics, including examples and best practices, see the Summary Metrics Guide.
Detailed quality checking results
Detailed row-level quality check results are added as additional columns to the output or quarantine (if defined) DataFrame or tables (if saved). These columns capture the outcomes of the checks performed on the input data.
The result columns are named _error and _warning by default, but you can customize them as described in the additional configuration section.
The result columns can be used to monitor and track data quality issues and for further processing, such as using in a dashboard, or other downstream applications.
Below is a sample output of a check stored in a result column (error or warning):
[
{
"name": "col_city_is_null",
"message": "Column 'city' is null",
"columns": ["city"],
"filter": "country = 'Poland'",
"function": "is_not_null",
"run_time": "2025-10-29T15:09:41.174Z",
"run_id": "2f9120cf-e9f2-446a-8278-12d508b00639"
"user_metadata": {"key1": "value1", "key2": "value2"},
},
]
The structure of the result columns is an array of struct containing the following fields (see the exact structure here):
name: name of the check (string type).message: message describing the quality issue (string type).columns: name of the column(s) where the quality issue was found (string type).filter: filter applied to if any (string type).function: rule/check function applied (string type).run_time: timestamp when the check was executed (timestamp type).run_id: unique run ID recorded in the detailed quality checking results as well as summary metrics to enable cross-referencing. When reusing the sameDQEngineand observer instances, the run ID stays the same. Each apply checks execution does not generate a new run ID for the same instance. It is only changed when new engine and observer (if using one) is created.user_metadata: optional key-value custom metadata provided by the user (dictionary type).
The below example demonstrates how to extract the results from a result columns in PySpark:
- Python
import pyspark.sql.functions as F
# apply quality checks
valid_df, invalid_df = dq_engine.apply_checks_and_split(input_df, checks)
# extract errors
results_df = invalid_df.select(
F.explode(F.col("_errors")).alias("result"),
).select(F.expr("result.*"))
# extract warnings
results_df = invalid_df.select(
F.explode(F.col("_warnings")).alias("result"),
).select(F.expr("result.*"))
results_df.show()
# If using summary metrics, you can drill down to detailed quality results
run_id = spark.table(metrics_table_name).collect()[0]["run_id"] # fetch any run_id as an example
filtered_results_df = results_df.filter(F.col("run_id") == run_id) # use run_id for filtering
filtered_results_df.show()
The results_df will contain the following columns:
+------------------+-----------------------+----------+--------------------+-------------+-------------------------------+--------------------------------------+----------------+
| name | message | columns | filter | function | run_time | run_id | user_metadata |
+------------------+-----------------------+----------+--------------------+-------------+-------------------------------+--------------------------------------+----------------+
| col_city_is_null | Column 'city' is null | ['city'] | country = 'Poland' | is_not_null | 2025-10-29T15:11:51.354+00:00 | 2f9120cf-e9f2-446a-8278-12d508b00639 | {} |
| ... | ... | ... | ... | ... | ... | ... | |
+------------------+-----------------------+----------+--------------------+-------------+-------------------------------+--------------------------------------+----------------+
An example of how to provide user metadata can be found in the Additional Configuration section.