Profiling and Quality Checks Generation
Data profiling can be run to profile input data and generate quality rule candidates with summary statistics.
The generated data quality rules (checks) candidates can be used as input for the quality checking as described here. In addition, the Lakeflow (DLT) generator can generate native Lakeflow Pipelines expectations.
You can run profiling and quality checks generation using the following approaches:
- Programmatic approach: you can use DQX classes to run the profiler and quality checks generator on DataFrames or tables, and save the generated checks to a storage.
- No-code approach (Profiler Workflow): you can run the profiler and quality checks generator on tables as a workflow if DQX is installed in the workspace as a tool. The workflow will save the generated checks to a file or table automatically.
See DQProfile Types for a complete list of profile types and associated DQX rules.
Data profiling is typically performed as a one-time action for the input dataset to discover the initial set of quality rule candidates. The check candidates should be manually reviewed before being applied to the data. This is not intended to be a continuously repeated or scheduled process, thereby also minimizing concerns regarding compute intensity and associated costs.
DQX offers AI-assisted generation of data quality rules based on user input in natural language. See more details in the AI-Assisted Data Quality Rules Generation section.
DQX also offers AI-assisted primary key detection to generate uniqueness rules. See more details in the AI-Assisted Primary Key Detection section.
Programmatic approach
You can use DQX classes to profile data and generate quality checks candidates programmatically. There are multiple storage options for saving the generated quality checks as described here.
Profiling a DataFrame
Data loaded as a DataFrame can be profiled using DQX classes to generate summary statistics and candidate data quality rules.
- Python
from databricks.labs.dqx.profiler.profiler import DQProfiler
from databricks.labs.dqx.profiler.generator import DQGenerator
from databricks.labs.dqx.profiler.dlt_generator import DQDltGenerator
from databricks.labs.dqx.config import WorkspaceFileChecksStorageConfig
from databricks.labs.dqx.engine import DQEngine
from databricks.sdk import WorkspaceClient
input_df = spark.read.table("catalog1.schema1.table1")
# profile input data
ws = WorkspaceClient()
profiler = DQProfiler(ws)
summary_stats, profiles = profiler.profile(input_df)
# generate DQX quality rules/checks
generator = DQGenerator(ws)
checks = generator.generate_dq_rules(profiles) # with default level "error"
dq_engine = DQEngine(ws)
# save checks as YAML in arbitrary workspace location
dq_engine.save_checks(checks, config=WorkspaceFileChecksStorageConfig(location="/Shared/App1/checks.yml"))
# generate Lakeflow Pipeline (DLT) expectations
dlt_generator = DQDltGenerator(ws)
dlt_expectations = dlt_generator.generate_dlt_rules(profiles, language="SQL")
print(dlt_expectations)
dlt_expectations = dlt_generator.generate_dlt_rules(profiles, language="Python")
print(dlt_expectations)
dlt_expectations = dlt_generator.generate_dlt_rules(profiles, language="Python_Dict")
print(dlt_expectations)
The profiler samples 30% of the data (sample ratio = 0.3) and limits the input to 1000 records by default. These and other configuration options can be customized as detailed in the Profiling Options section.
Profiling a table
Profiling and quality checks generation can be run on individual tables.
- Python
from databricks.labs.dqx.profiler.profiler import DQProfiler
from databricks.sdk import WorkspaceClient
from databricks.labs.dqx.config import InputConfig
ws = WorkspaceClient()
profiler = DQProfiler(ws)
summary_stats, profiles = profiler.profile_table(
input_config=InputConfig(location="catalog1.schema1.table1")
columns=["col1", "col2", "col3"], # specify columns to profile
)
print("Summary Statistics:", summary_stats)
print("Generated Profiles:", profiles)
Profiling multiple tables
The profiler can discover and profile multiple tables accessible via Unity Catalog. Tables can be passed explicitly as a list or be included/excluded using wildcard patterns (e.g. "catalog.schema.*").
- Python
from databricks.labs.dqx.profiler.profiler import DQProfiler
from databricks.sdk import WorkspaceClient
ws = WorkspaceClient()
profiler = DQProfiler(ws)
# Profile several tables by name
results = profiler.profile_tables_for_patterns(
patterns=["main.data.table_001", "main.data.table_002"]
)
# Process results for each table
for table, (summary_stats, profiles) in results.items():
print(f"Table: {table}")
print(f"Table statistics: {summary_stats}")
print(f"Generated profiles: {profiles}")
# Profile several tables by wildcard patterns
results = profiler.profile_tables_for_patterns(
patterns=["main.*", "data.*"]
)
# Process results for each table
for table, (summary_stats, profiles) in results.items():
print(f"Table: {table}")
print(f"Table statistics: {summary_stats}")
print(f"Generated profiles: {profiles}")
# Exclude tables matching specific patterns
results = profiler.profile_tables_for_patterns(
patterns=["sys.*", "*_tmp"],
exclude_matched=True
)
# Process results for each table
for table, (summary_stats, profiles) in results.items():
print(f"Table: {table}")
print(f"Table statistics: {summary_stats}")
print(f"Generated profiles: {profiles}")
# Exclude tables matching specific patterns
results = profiler.profile_tables_for_patterns(
patterns=["sys.*", "*_tmp"],
exclude_patterns=["*_dq_output", "*_dq_quarantine"], # skip existing output tables using suffixes
)
# Process results for each table
for table, (summary_stats, profiles) in results.items():
print(f"Table: {table}")
print(f"Table statistics: {summary_stats}")
print(f"Generated profiles: {profiles}")
Profiling and expectations generation for Lakeflow Pipelines
You can integrate DQX quality checking directly with Lakeflow Pipelines (formerly Delta Live Tables, DLT). You can also generate Lakeflow Pipelines expectation statements from profiler results.
- Python
from databricks.labs.dqx.profiler.dlt_generator import DQDltGenerator
from databricks.sdk import WorkspaceClient
# After profiling your data
ws = WorkspaceClient()
profiler = DQProfiler(ws)
summary_stats, profiles = profiler.profile(input_df)
# Generate Lakeflow Pipelines expectations
dlt_generator = DQDltGenerator(ws)
# Generate SQL expectations
sql_expectations = dlt_generator.generate_dlt_rules(profiles, language="SQL")
print("SQL Expectations:")
for expectation in sql_expectations:
print(expectation)
# Output example:
# CONSTRAINT user_id_is_null EXPECT (user_id is not null)
# CONSTRAINT age_isnt_in_range EXPECT (age >= 18 and age <= 120)
# Generate SQL expectations with actions
sql_with_drop = dlt_generator.generate_dlt_rules(profiles, language="SQL", action="drop")
print("SQL Expectations with DROP action:")
for expectation in sql_with_drop:
print(expectation)
# Output example:
# CONSTRAINT user_id_is_null EXPECT (user_id is not null) ON VIOLATION DROP ROW
# Generate Python expectations
python_expectations = dlt_generator.generate_dlt_rules(profiles, language="Python")
print("Python Expectations:")
print(python_expectations)
# Output example:
# @dlt.expect_all({
# "user_id_is_null": "user_id is not null",
# "age_isnt_in_range": "age >= 18 and age <= 120"
# })
# Generate Python dictionary format
dict_expectations = dlt_generator.generate_dlt_rules(profiles, language="Python_Dict")
print("Python Dictionary Expectations:")
print(dict_expectations)
# Output example:
# {
# "user_id_is_null": "user_id is not null",
# "age_isnt_in_range": "age >= 18 and age <= 120"
# }
No-code approach (Profiler Workflow)
You can run profiler workflow to profile tables and generate quality checks candidates. The profiler workflow saves the generated checks automatically in the checks location as defined in the configuration file.
You need to install DQX as a tool in the workspace to have the profiler workflow available (see installation guide). The workflow is not scheduled to run automatically by default, minimizing concerns regarding compute and associated costs. Profiling is typically done as a one-time action for the input dataset to discover the initial set of quality rule candidates.
You can run the profiler workflow from Databricks UI or by using Databricks CLI:
# Run for all run configs in the configuration file
databricks labs dqx profile
# Run for a specific run config in the configuration file
databricks labs dqx profile --run-config "default"
# Run for all tables/views matching wildcard patterns. Conventions:
# * Run config from configuration file is used as a template for all relevant fields except location
# * Input table location is derived from the patterns
# * For table-based checks location, checks are saved to the specified table
# * For file-based checks location, file in the path is replaced with <input_table>.yml. In addition, if the location is specified as a relative path, it is prefixed with the installation folder
# * By default, output and quarantine tables are excluded based on suffixes
# * Default for output table suffix is "_dq_output"
# * Default for quarantine table suffix is "_dq_quarantine"
databricks labs dqx profile --run-config "default" --patterns "main.product001.*;main.product002"
# Run for wildcard patterns and exclude patterns
databricks labs dqx profile --run-config "default" --patterns "main.product001.*;main.product002" --exclude-patterns "*_output;*_quarantine"
You can also provide --timeout-minutes option.
When running the profiler workflow using Databricks API or UI, you have the same execution options:
- By default, the workflow executes across all defined run configs within the configuration file.
- To execute the workflow for a specific run config, specify the
run_config_nameparameter during execution. - To execute the workflow for tables/views matching wildcard patterns, provide value for
run_config_nameandpatternsparameters during execution. The following parameters are supported for pattern based execution:patterns: Accepts a semicolon-delimited list of patterns, e.g., "catalog.schema1.*;catalog.schema2.table1".run_config_name: Specifies the run config to use as a template for all relevant settings, except location field ('input_config.location') which is derived from the patterns.- The output and quarantine table names are derived by appending the value of
output_table_suffixandquarantine_table_suffixjob parameters, respectively, to the input table name. - If the
checks_locationin the run config points to a table, the checks will be saved to that table. If thechecks_locationin the run config points to a file, file name is replaced with "<input_table>.yml". In addition, if the location is specified as a relative path, it is prefixed with the workspace installation folder. For example:- If "checks_location=catalog.schema.table", the location will be resolved to "catalog.schema.table" or "database.schema.table" in case of using Lakebase to store checks.
- If "checks_location=folder/checks.yml", the location will be resolved to "install_folder/folder/<input_table>.yml".
- If "checks_location=/App/checks.yml", the location will be resolved to "/App/<input_table>.yml".
- If "checks_location=/Volume/catalog/schema/folder/checks.yml", the location will be resolved to "/Volume/catalog/schema/folder/<input_table>.yml".
exclude_patterns: (optional) Accepts a semicolon-delimited list of patterns to exclude, e.g., "*_output;*_quarantine".- By default, the workflow skips output and quarantine tables automatically based on the
output_table_suffixandquarantine_table_suffixjob parameters. This prevents potential profiling of the output and quarantine tables generated by the quality checker workflow. To disable this behavior, clear the values ofoutput_table_suffixandquarantine_table_suffixin the job parameters. output_table_suffix: (optional) Suffix used for the output table names (default: "_dq_output").quarantine_table_suffix: (optional) Suffix used for to the quarantine table names (default: "_dq_quarantine").
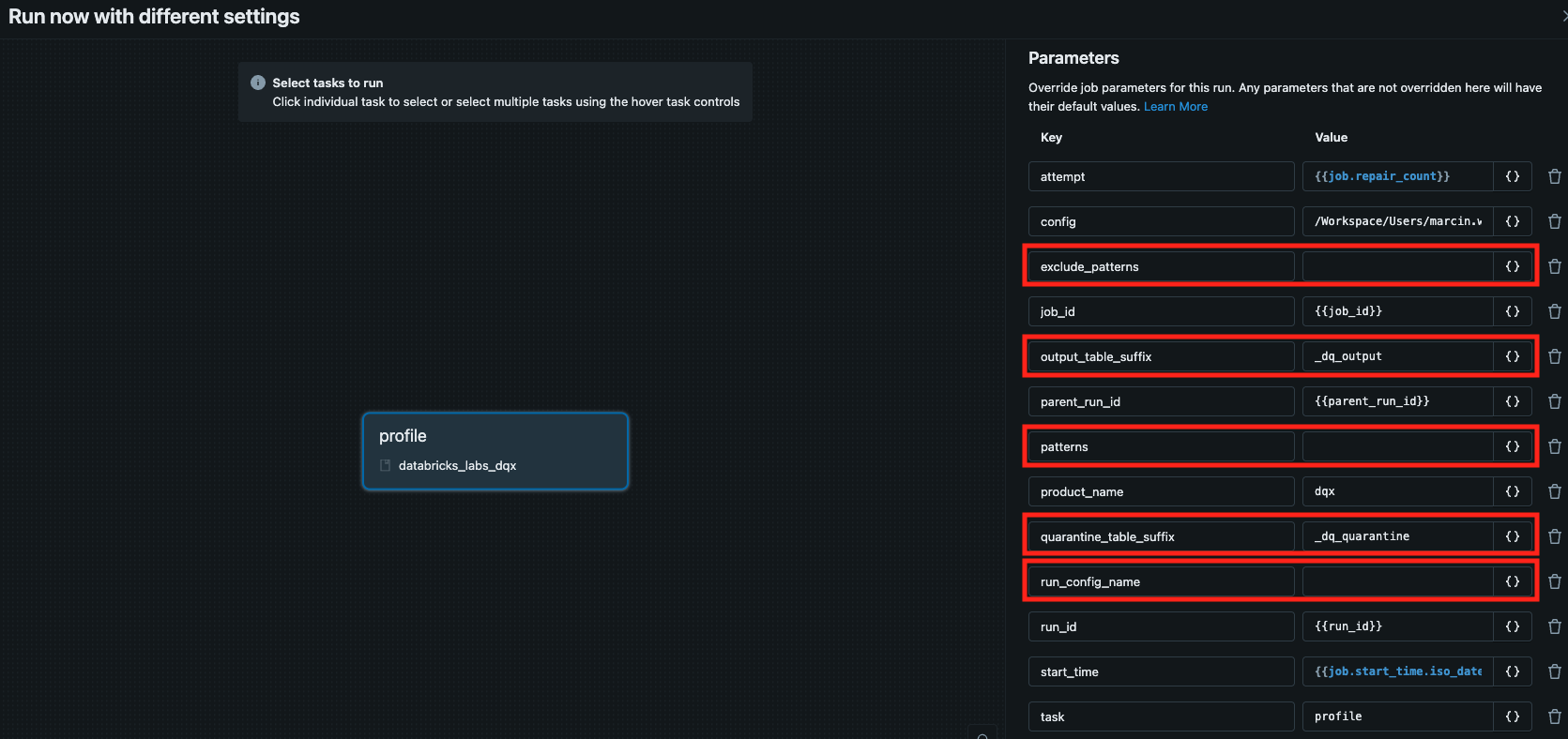
See the relevant params in red:
Execution logs from the profiler are printed in the console and saved in the installation folder. You can display the logs from the latest profiler workflow run by executing:
databricks labs dqx logs --workflow profiler
The generated quality rule candidates and summary statistics will be saved in the checks_location set in the run config of the configuration file.
You can open the configuration file (.dqx/config.yml) in the installation folder from Databricks UI or by executing the following Databricks CLI command:
databricks labs dqx open-remote-config
The following fields from the configuration file are used by the profiler workflow:
input_configconfiguration for the input data. 'location' is autogenerated when the workflow is executed for patterns.checks_location: location of the quality checks in storage. Autogenerated when the workflow is executed for patterns.profiler_config: configuration for the profiler containing:summary_stats_file: relative location within the installation folder of the summary statistics (default: 'profile_summary.yml')sample_fraction: fraction of data to sample for profiling.sample_seed: seed for reproducible sampling.limit: maximum number of records to analyze.filter: filter for the input data as a string SQL expression
checks_user_requirements: (optional) user input for AI-assisted rule generation. Only supported ifserverless_clustersis enabled.llm_config: (optional) configuration for the llm-assisted featuresserverless_clusters: whether to use serverless clusters for running the workflow (default: 'true'). Using serverless clusters is recommended as it allows for automated cluster management and scaling.profiler_spark_conf: (optional) spark configuration to use for the profiler workflow, only applicable ifserverless_clustersis set to 'false'.profiler_override_clusters: (optional) cluster configuration to use for profiler workflow, only applicable ifserverless_clustersis set to 'false'.profiler_max_parallelism: (optional) max parallelism for profiling multiple tables (default: 4)
Example of the configuration file (relevant fields only):
serverless_clusters: true # default is true, set to false to use standard clusters
profiler_max_parallelism: 4 # max parallelism for profiling multiple tables
llm_config: # configuration for the llm-assisted features
model:
model_name: "databricks/databricks-claude-sonnet-4-5"
api_key: xxx # optional API key for the model as secret in the format: secret_scope/secret_key. Not required by foundational models
api_base: xxx # optional API base for the model as secret in the format: secret_scope/secret_key. Not required by foundational models
run_configs:
- name: default
checks_location: catalog.table.checks # can be a table or file
input_config:
format: delta
location: /databricks-datasets/delta-sharing/samples/nyctaxi_2019
is_streaming: false # set to true if wanting to run it using streaming
profiler_config:
limit: 1000
sample_fraction: 0.3
summary_stats_file: profile_summary_stats.yml
filter: "maintenance_type = 'preventive'"
llm_primary_key_detection: false
checks_user_requirements: "business rules description" # optional user input for the llm-assisted features
Profiling options
The profiler supports extensive configuration options to customize the profiling behavior.
Profiling options for a single table
- Python
- Workflows
For code-level integration you can use options parameter to pass a dictionary with custom options when profiling a DataFrame or table.
from databricks.labs.dqx.profiler.profiler import DQProfiler
from databricks.sdk import WorkspaceClient
from databricks.labs.dqx.config import InputConfig
# Custom profiling options
custom_options = {
# Sampling options
"sample_fraction": 0.2, # Sample 20% of the data
"sample_seed": 42, # Seed for reproducible sampling
"limit": 2000, # Limit to 2000 records after sampling
# Outlier detection options
"remove_outliers": True, # Enable outlier detection for min/max rules
"outlier_columns": ["price", "age"], # Only detect outliers in specific columns
"num_sigmas": 2.5, # Use 2.5 standard deviations for outlier detection
# Null value handling
"max_null_ratio": 0.05, # Generate is_not_null rule if <5% nulls
# String handling
"trim_strings": True, # Trim whitespace from strings before analysis
"max_empty_ratio": 0.02, # Generate is_not_null_or_empty rule if <2% empty strings
# Distinct value analysis
"distinct_ratio": 0.01, # Generate is_in rule if <1% distinct values
"max_in_count": 20, # Maximum items in is_in rule list
# Value rounding
"round": True, # Round min/max values for cleaner rules
# Filter Options
"filter": None, # No filter (SQL expression)
"llm_primary_key_detection": True, # Detect PKs and generate uniqueness rule if found
}
ws = WorkspaceClient()
profiler = DQProfiler(ws)
# Apply custom options for profiling a DataFrame
summary_stats, profiles = profiler.profile(input_df, options=custom_options)
# Apply custom options for profiling a table
summary_stats, profiles = profiler.profile_table(
input_config=InputConfig(location="catalog1.schema1.table1"),
columns=["col1", "col2", "col3"],
options=custom_options
)
For the profiler workflow, you can set profiling options for each run config in the configuration file using the following fields:
profiler_config: configuration for the profiler containing:summary_stats_file: relative location within the installation folder of the summary statistics (default: 'profile_summary.yml')sample_fraction: fraction of data to sample for profiling.sample_seed: seed for reproducible sampling.limit: maximum number of records to analyze.filter: string SQL expression to filter the input data
You can open the configuration file to check available run configs and adjust the settings if needed:
databricks labs dqx open-remote-config
Example:
run_configs:
- name: default
profiler_config:
sample_fraction: 0.3 # Fraction of data to sample (30%)
limit: 1000 # Maximum number of records to analyze
sample_seed: 42 # Seed for reproducible results
llm_primary_key_detection: false # whether to use llm-assisted pk detection to generate uniqueness check
...
More detailed options such as 'num_sigmas' are not configurable when using profiler workflow. They are only available for programmatic integration.
Profiling options for multiple tables
When profiling multiple tables, you can pass a list of dictionaries to apply different options to each one. Wildcard patterns are supported, allowing you to match table names and apply specific options based on those patterns.
- Python
from databricks.labs.dqx.profiler.profiler import DQProfiler
from databricks.sdk import WorkspaceClient
ws = WorkspaceClient()
profiler = DQProfiler(ws)
tables = [
"dqx.bronze.table_001",
"dqx.silver.table_001",
"dqx.silver.table_002",
]
# Custom options with wildcard patterns
custom_table_options = [
{
"table": "*", # matches all tables by pattern
"options": {"sample_fraction": 0.5}
},
{
"table": "dqx.silver.*", # matches tables in the 'dqx.silver' schema by pattern
"options": {"num_sigmas": 5}
},
{
"table": "dqx.silver.table_*", # matches tables in 'dqx.silver' schema and having 'table_' prefix
"options": {"num_sigmas": 5}
},
{
"table": "dqx.silver.table_002", # matches a specific table, overrides generic option
"options": {"sample_fraction": 0.1}
},
]
# Profile multiple tables using custom options
results = profiler.profile_tables_for_patterns(patterns=tables, options=custom_table_options)
# Profile multiple tables by wildcard patterns using custom options
results = profiler.profile_tables_for_patterns(
patterns=["dqx.*"],
options=custom_table_options
)
Understanding summary statistics and profiles
To ensure efficiency, summary statistics are computed on a sampled subset of your data, not the entire dataset. By default, the profiler proceeds as follows:
- It samples the rows of the dataset you provide by using Spark’s
DataFrame.sample(defaultsample_fraction = 0.3), which is probabilistic, hence the sampled row count is not guaranteed to be exact; the number may be slightly higher or lower. No filters are applied by default. - A limit is applied to the sampled rows (default
limit = 1000). - Statistics are computed on this final sampled-and-limited DataFrame.
These are defaults that can be customized programmatically as detailed in the Profiling Options section, or via profiler_config in the configuration file if DQX is installed as a tool.
Rule of thumb: count ≈ min(limit, sample_fraction × total_rows).
Summary Statistics Reference
| Field | Meaning | Notes |
|---|---|---|
count | Rows actually profiled (after sampling and limit) | ≈ min(limit, sample_fraction × total_rows) |
mean | Arithmetic average of non-null numeric values | N/A for non-numeric |
stddev | Sample standard deviation of non-null numeric values | N/A for non-numeric |
min | Smallest non-null value | String = lexicographic; Date/Timestamp = earliest; Numeric = minimum |
25 / 50 / 75 | Approximate 25th/50th/75th percentiles of non-null numeric values | Uses Spark approximate quantiles |
max | Largest non-null value | String = lexicographic; Date/Timestamp = latest; Numeric = maximum |
count_non_null | Number of non-null entries within the profiled rows | |
count_null | Number of null entries within the profiled rows | count_non_null + count_null = count |
Performance considerations
When profiling large datasets, use sampling and limits for best performance.
- Python
- Workflows
# For large datasets, use aggressive sampling
large_dataset_opts = {
"sample_fraction": 0.01, # Sample only 1% for very large datasets
"limit": 10000, # Increase limit for better statistical accuracy
"sample_seed": 42, # Use consistent seed for reproducible results
}
# For medium datasets, use moderate sampling
medium_dataset_opts = {
"sample_fraction": 0.1, # Sample 10%
"limit": 5000, # Reasonable limit
}
# For small datasets, disable sampling
small_dataset_opts = {
"sample_fraction": None, # Use all data
"limit": None, # No limit
}
Use the following configuration in the configuration file to configure the profiler workflow:
For large datasets, use aggressive sampling:
profiler_config:
sample_fraction: 0.01 # Sample only 1% for very large datasets
limit: 10000 # Increase limit for better statistical accuracy
sample_seed: 42 # Use consistent seed for reproducible results
For medium datasets, use moderate sampling:
profiler_config:
sample_fraction: 0.1 # Sample 10%
limit: 5000 # Reasonable limit
For small datasets, disable sampling:
profiler_config:
sample_fraction: null # Use all data
limit: null # No limit
Summary statistics from limited samples may not reflect the characteristics of the overall dataset. Balance the sampling rate and limits with your desired profile accuracy. Manually review and tune rules generated from profiles on sample data to ensure correctness.