Rasterio UDFs
Intro
Rasterio (https://rasterio.readthedocs.io/en/latest/) is a Python library for reading and writing geospatial raster datasets. It uses GDAL (https://gdal.org/) for file I/O and raster formatting and provides a Python API for GDAL functions. It is a great library for working with raster data in Python and it is a popular choice for many geospatial data scientists. Rasterio UDFs provide a way to use Rasterio Python API in Spark for distributed processing of raster data. The data structures used by Mosaic are compatible with Rasterio and can be used interchangeably. In this section we will show how to use Rasterio UDFs to process raster data in Mosaic + Spark. We assume that you have a basic understanding of Rasterio and GDAL.
Please note that we advise the users to set these configuration to ensure proper distribution.
spark.conf.set("spark.sql.execution.arrow.enabled", "true")
spark.conf.set("spark.sql.execution.arrow.maxRecordsPerBatch", "1024")
spark.conf.set("spark.sql.execution.arrow.fallback.enabled", "true")
spark.conf.set("spark.sql.adaptive.coalescePartitions.enabled", "false")
spark.conf.set("spark.sql.shuffle.partitions", "400")
Rasterio raster plotting
In this example we will show how to plot a raster file using Rasterio Python API.
Firstly we will create a spark DataFrame from a directory of raster files.
df = spark.read.format("gdal").load("dbfs:/path/to/raster/files").repartition(400)
df.show()
+-----------------------------------------------------------+------------------------------+-----------+---------------------+-------+-------+-----------+----------------------+-------------+-------+---------------------------------------------------------------------------------------------------------------+
| path | modificationTime | length | uuid | ySize | xSize | bandCount | metadata | subdatasets | srid | tile |
+-----------------------------------------------------------+------------------------------+-----------+---------------------+-------+-------+-----------+----------------------+-------------+-------+---------------------------------------------------------------------------------------------------------------+
| dbfs:/FileStore/geospatial/odin/alaska/B02/-424495268.tif | 1970-01-20T15:49:53.135+0000 | 211660514 | 7836235824828840960 | 10980 | 10980 | 1 | {AREA_OR_POINT=Po... | {} | 32602 | {index_id: 593308294097928191, raster: [00 01 10 ... 00], parentPath: "dbfs:/path_to_file", driver: "GTiff" } |
| dbfs:/FileStore/geospatial/odin/alaska/B02/-524425268.tif | 1970-01-20T15:49:53.135+0000 | 212060218 | 7836235824828840961 | 10980 | 10980 | 1 | {AREA_OR_POINT=Po... | {} | 32602 | {index_id: 593308294097927192, raster: [00 01 10 ... 00], parentPath: "dbfs:/path_to_file", driver: "GTiff" } |
| dbfs:/FileStore/geospatial/odin/alaska/B02/1241323268.tif | 1970-01-20T15:49:53.135+0000 | 211660897 | 7836235824828840962 | 10980 | 10980 | 1 | {AREA_OR_POINT=Po... | {} | 32602 | {index_id: 593308294097929991, raster: [00 01 10 ... 00], parentPath: "dbfs:/path_to_file", driver: "GTiff" } |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
+-----------------------------------------------------------+------------------------------+-----------+---------------------+-------+-------+-----------+----------------------+-------------+-------+---------------------------------------------------------------------------------------------------------------+
Next we will define a function that will plot a given raster file.
import numpy as np
import rasterio
from rasterio.io import MemoryFile
from io import BytesIO
from pyspark.sql.functions import udf
def plot_raster(raster):
fig, ax = pyplot.subplots(1, figsize=(12, 12))
with MemoryFile(BytesIO(raster)) as memfile:
with memfile.open() as src:
show(src, ax=ax)
pyplot.show()
Finally we will apply the function to the DataFrame collected results. Note that in order to plot the raster we need to collect the results to the driver. Please apply reasonable filters to the DataFrame before collecting the results.
plot_raster(df.select("tile").limit(1).collect()[0]["tile"]["raster"])
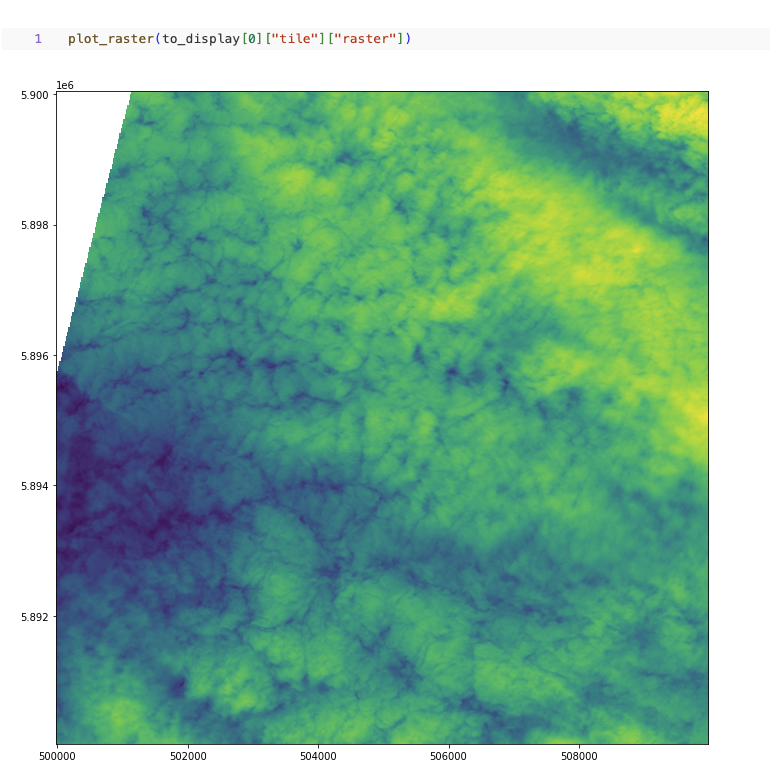
Fig 1. Plot raster using Rasterio Python API
UDF example for computing band statistics
In this example we will show how to compute band statistics for a raster file.
Firstly we will create a spark DataFrame from a directory of raster files.
df = spark.read.format("gdal").load("dbfs:/path/to/raster/files").repartition(400)
df.show()
+-----------------------------------------------------------+------------------------------+-----------+---------------------+-------+-------+-----------+----------------------+-------------+-------+---------------------------------------------------------------------------------------------------------------+
| path | modificationTime | length | uuid | ySize | xSize | bandCount | metadata | subdatasets | srid | tile |
+-----------------------------------------------------------+------------------------------+-----------+---------------------+-------+-------+-----------+----------------------+-------------+-------+---------------------------------------------------------------------------------------------------------------+
| dbfs:/FileStore/geospatial/odin/alaska/B02/-424495268.tif | 1970-01-20T15:49:53.135+0000 | 211660514 | 7836235824828840960 | 10980 | 10980 | 1 | {AREA_OR_POINT=Po... | {} | 32602 | {index_id: 593308294097928191, raster: [00 01 10 ... 00], parentPath: "dbfs:/path_to_file", driver: "GTiff" } |
| dbfs:/FileStore/geospatial/odin/alaska/B02/-524425268.tif | 1970-01-20T15:49:53.135+0000 | 212060218 | 7836235824828840961 | 10980 | 10980 | 1 | {AREA_OR_POINT=Po... | {} | 32602 | {index_id: 593308294097927192, raster: [00 01 10 ... 00], parentPath: "dbfs:/path_to_file", driver: "GTiff" } |
| dbfs:/FileStore/geospatial/odin/alaska/B02/1241323268.tif | 1970-01-20T15:49:53.135+0000 | 211660897 | 7836235824828840962 | 10980 | 10980 | 1 | {AREA_OR_POINT=Po... | {} | 32602 | {index_id: 593308294097929991, raster: [00 01 10 ... 00], parentPath: "dbfs:/path_to_file", driver: "GTiff" } |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
+-----------------------------------------------------------+------------------------------+-----------+---------------------+-------+-------+-----------+----------------------+-------------+-------+---------------------------------------------------------------------------------------------------------------+
Next we will define a function that will compute band statistics for a given raster file.
import numpy as np
import rasterio
from rasterio.io import MemoryFile
from io import BytesIO
from pyspark.sql.functions import udf
@udf("double")
def compute_band_mean(raster):
with MemoryFile(BytesIO(raster)) as memfile:
with memfile.open() as dataset:
return dataset.statistics(bidx = 1).mean
Finally we will apply the function to the DataFrame.
df.select(compute_band_mean("tile.raster")).show()
+---------------------------+
| compute_band_mean(raster) |
+---------------------------+
| 0.0111000000000000|
| 0.0021000000000000|
| 0.3001000000000000|
| ... |
+---------------------------+
UDF example for computing NDVI
In this example we will show how to compute NDVI for a raster file. NDVI is a common index used to assess vegetation health. It is computed as follows: ndvi = (nir - red) / (nir + red). NDVI output is a single band raster file with values in the range [-1, 1]. We will show how to return a raster object as a result of a UDF.
Firstly we will create a spark DataFrame from a directory of raster files.
df = spark.read.format("gdal").load("dbfs:/path/to/raster/files").repartition(400)
df.show()
+-----------------------------------------------------------+------------------------------+-----------+---------------------+-------+-------+-----------+----------------------+-------------+-------+---------------------------------------------------------------------------------------------------------------+
| path | modificationTime | length | uuid | ySize | xSize | bandCount | metadata | subdatasets | srid | tile |
+-----------------------------------------------------------+------------------------------+-----------+---------------------+-------+-------+-----------+----------------------+-------------+-------+---------------------------------------------------------------------------------------------------------------+
| dbfs:/FileStore/geospatial/odin/alaska/B02/-424495268.tif | 1970-01-20T15:49:53.135+0000 | 211660514 | 7836235824828840960 | 10980 | 10980 | 1 | {AREA_OR_POINT=Po... | {} | 32602 | {index_id: 593308294097928191, raster: [00 01 10 ... 00], parentPath: "dbfs:/path_to_file", driver: "GTiff" } |
| dbfs:/FileStore/geospatial/odin/alaska/B02/-524425268.tif | 1970-01-20T15:49:53.135+0000 | 212060218 | 7836235824828840961 | 10980 | 10980 | 1 | {AREA_OR_POINT=Po... | {} | 32602 | {index_id: 593308294097927192, raster: [00 01 10 ... 00], parentPath: "dbfs:/path_to_file", driver: "GTiff" } |
| dbfs:/FileStore/geospatial/odin/alaska/B02/1241323268.tif | 1970-01-20T15:49:53.135+0000 | 211660897 | 7836235824828840962 | 10980 | 10980 | 1 | {AREA_OR_POINT=Po... | {} | 32602 | {index_id: 593308294097929991, raster: [00 01 10 ... 00], parentPath: "dbfs:/path_to_file", driver: "GTiff" } |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
+-----------------------------------------------------------+------------------------------+-----------+---------------------+-------+-------+-----------+----------------------+-------------+-------+---------------------------------------------------------------------------------------------------------------+
Next we will define a function that will compute NDVI for a given raster file.
import numpy as np
import rasterio
from rasterio.io import MemoryFile
from io import BytesIO
from pyspark.sql.functions import udf
@udf("binary")
def compute_ndvi(raster, nir_band, red_band):
with MemoryFile(BytesIO(raster)) as memfile:
with memfile.open() as dataset:
red = dataset.read(red_band)
nir = dataset.read(nir_band)
ndvi = (nir - red) / (nir + red)
profile = dataset.profile
profile.update(count = 1, dtype = rasterio.float32)
# Write the NDVI to a tmp file and return it as binary
# This is a workaround an issue occurring when using
# MemoryFile for writing using an updated profile
with tempfile.NamedTemporaryFile() as tmp:
with rasterio.open(tmp.name, "w", **profile) as dst:
dst.write(ndvi.astype(rasterio.float32))
with open(tmp.name, "rb") as f:
return f.read()
Finally we will apply the function to the DataFrame.
df.select(compute_ndvi("tile.raster", lit(1), lit(2))).show()
# The output is a binary column containing the NDVI raster
+------------------------------+
| compute_ndvi(raster, 1, 2) |
+------------------------------+
| 000000 ... 00000000000000000 |
| 000000 ... 00000000000000000 |
| 000000 ... 00000000000000000 |
| ... |
+------------------------------+
# We can update the tile column with the NDVI raster in place as well
# This will overwrite the existing raster field in the tile column
df.select(col("tile").withField("raster", compute_ndvi("tile.raster", lit(1), lit(2)))).show()
+-----------------------------------------------------------+------------------------------+-----------+---------------------+-------+-------+-----------+----------------------+-------------+-------+---------------------------------------------------------------------------------------------------------------+
| path | modificationTime | length | uuid | ySize | xSize | bandCount | metadata | subdatasets | srid | tile |
+-----------------------------------------------------------+------------------------------+-----------+---------------------+-------+-------+-----------+----------------------+-------------+-------+---------------------------------------------------------------------------------------------------------------+
| dbfs:/FileStore/geospatial/odin/alaska/B02/-424495268.tif | 1970-01-20T15:49:53.135+0000 | 211660514 | 7836235824828840960 | 10980 | 10980 | 1 | {AREA_OR_POINT=Po... | {} | 32602 | {index_id: 593308294097928191, raster: [00 01 10 ... 00], parentPath: "dbfs:/path_to_file", driver: "GTiff" } |
| dbfs:/FileStore/geospatial/odin/alaska/B02/-524425268.tif | 1970-01-20T15:49:53.135+0000 | 212060218 | 7836235824828840961 | 10980 | 10980 | 1 | {AREA_OR_POINT=Po... | {} | 32602 | {index_id: 593308294097927192, raster: [00 01 10 ... 00], parentPath: "dbfs:/path_to_file", driver: "GTiff" } |
| dbfs:/FileStore/geospatial/odin/alaska/B02/1241323268.tif | 1970-01-20T15:49:53.135+0000 | 211660897 | 7836235824828840962 | 10980 | 10980 | 1 | {AREA_OR_POINT=Po... | {} | 32602 | {index_id: 593308294097929991, raster: [00 01 10 ... 00], parentPath: "dbfs:/path_to_file", driver: "GTiff" } |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
+-----------------------------------------------------------+------------------------------+-----------+---------------------+-------+-------+-----------+----------------------+-------------+-------+---------------------------------------------------------------------------------------------------------------+
UDF example for writing raster files to disk
In this example we will show how to write a raster file to disk using Rasterio Python API. This is an examples showing how to materialize a raster binary object as a raster file on disk. The format of the output file should match the driver format of the binary object.
Firstly we will create a spark DataFrame from a directory of raster files.
df = spark.read.format("gdal").load("dbfs:/path/to/raster/files").repartition(400)
df.show()
+-----------------------------------------------------------+------------------------------+-----------+---------------------+-------+-------+-----------+----------------------+-------------+-------+---------------------------------------------------------------------------------------------------------------+
| path | modificationTime | length | uuid | ySize | xSize | bandCount | metadata | subdatasets | srid | tile |
+-----------------------------------------------------------+------------------------------+-----------+---------------------+-------+-------+-----------+----------------------+-------------+-------+---------------------------------------------------------------------------------------------------------------+
| dbfs:/FileStore/geospatial/odin/alaska/B02/-424495268.tif | 1970-01-20T15:49:53.135+0000 | 211660514 | 7836235824828840960 | 10980 | 10980 | 1 | {AREA_OR_POINT=Po... | {} | 32602 | {index_id: 593308294097928191, raster: [00 01 10 ... 00], parentPath: "dbfs:/path_to_file", driver: "GTiff" } |
| dbfs:/FileStore/geospatial/odin/alaska/B02/-524425268.tif | 1970-01-20T15:49:53.135+0000 | 212060218 | 7836235824828840961 | 10980 | 10980 | 1 | {AREA_OR_POINT=Po... | {} | 32602 | {index_id: 593308294097927192, raster: [00 01 10 ... 00], parentPath: "dbfs:/path_to_file", driver: "GTiff" } |
| dbfs:/FileStore/geospatial/odin/alaska/B02/1241323268.tif | 1970-01-20T15:49:53.135+0000 | 211660897 | 7836235824828840962 | 10980 | 10980 | 1 | {AREA_OR_POINT=Po... | {} | 32602 | {index_id: 593308294097929991, raster: [00 01 10 ... 00], parentPath: "dbfs:/path_to_file", driver: "GTiff" } |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
+-----------------------------------------------------------+------------------------------+-----------+---------------------+-------+-------+-----------+----------------------+-------------+-------+---------------------------------------------------------------------------------------------------------------+
Next we will define a function that will write a given raster file to disk.
import numpy as np
import rasterio
from rasterio.io import MemoryFile
from io import BytesIO
from pyspark.sql.functions import udf
from pathlib import Path
@udf("string")
def write_raster(raster, file_id, parent_dir):
with MemoryFile(BytesIO(raster)) as memfile:
with memfile.open() as dataset:
Path(outputpath).mkdir(parents=True, exist_ok=True)
extensions_map = rasterio.drivers.raster_driver_extensions()
driver_map = {v: k for k, v in extensions_map.items()}
extension = driver_map[dataset.driver]
path = f"{parent_dir}/{file_id}.{extension}"
# If you want to write the raster to a different format
# you can update the profile here. Note that the extension
# should match the driver format
with rasterio.open(path, "w", **dataset.profile) as dst:
dst.write(dataset.read())
return path
Finally we will apply the function to the DataFrame.
df.select(write_raster("tile.raster", "uuid", lit("dbfs:/path/to/output/dir"))).show()
+-------------------------------------+
| write_raster(raster, output, output)|
+-------------------------------------+
| dbfs:/path/to/output/dir/1234.tif |
| dbfs:/path/to/output/dir/4545.tif |
| dbfs:/path/to/output/dir/3215.tif |
| ... |
+-------------------------------------+